Collaborations
GUDMAP, An NIH Project.
It aims to build the GUDMAP morphological atlas and the GUDMAP in situ and micro-array gene expression database to facilitate our understanding of genitourinary (GU) development and disease. The GUDMAP Atlas will be linked to the gene expression database, cell catalogue and other relevant resources. Together they provide a platform for modeling the processes of development, disease, and therapy.
EuReGene, A European Project.
An estimated 4.5 Million Europeans suffer from renal disorders. The death rate in patients with renal failure is 20% annually. The goal of this project is to discover genes responsible for renal development and disease, their proteins and their actions. To achieve this goal, a consortium of leading scientists, clinicians and SME partners will focus on the development of novel technologies and discovery tools in functional genomics and their application to kidney research.

EURExpress, A EU project funded under the VI Framework
It proposes a transcriptome-wide acquisition of expression patterns chiefly by means of in situ hybridization (ISH) with non-radioactive probes. More than 20,000 genes by RNA in situ hybridization on sagittal sections from E14.5 wild type murine embryos will be collected. The data will form a web-linked, interactive digital transcriptome atlas of embryonic mouse.
DGEMap stands for Developmental Gene Expression Map.

It is the first ever "Design Study" for a pan-European research infrastructure dedicated to the analysis of gene expression patterns in early human development.
Newcastle University (UK) was granted the coordination of DGEMap by the European Commission under the Sixth Framework Programme.
The project started in April 2005 and will end in July 2008.
Institute of Human Genetics, University of Newcastle.
Human Brain Gene Expression Atlas Project is aimed at creating 3-D reconstructions of brains at different developmental stages. The reconstructions will include anatomical information and be used to map gene expression data which together will build into an interactive database.
Dr Martin Ringwald, Jackson Laboratory, USA.

Dr Martin Ringwald and the GXD programme within the division of informatics at the Jackson Laboratory has collaborated with emap from the beginning of the program with the goal of providing complementary and interoperable gene-expression resources for the mouse community. The emap mouse nomenclature database has been incorporated in the GXD gene-expression database.
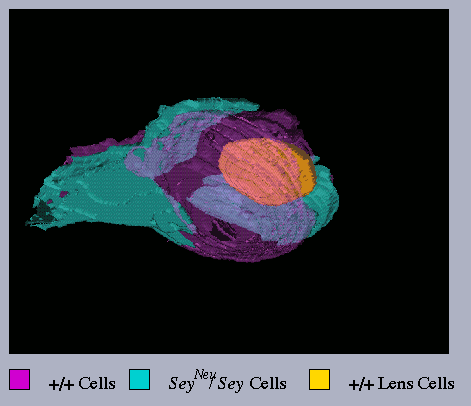
Dr Jane Quinn & Dr John West, Department of Obstetrics, University of Edinburgh.
Investigating effects of Pax6 on cell mixing in SeyNeu/Sey <-> +/+ chimaeric fetal mouse eyes using 3-dimensional reconstruction techniques.
Department of Anatomy, University of Edinburgh.
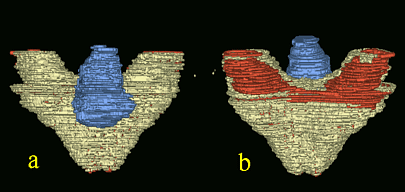
Analysis of fused maxillary dentition in p53-deficient exencaphalic mice.
Glaxo Welcome Medicines Research Centre, UK.
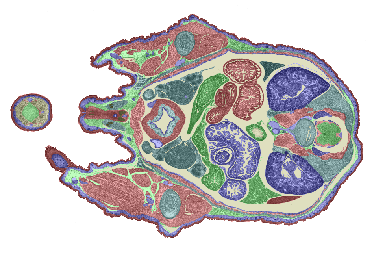
Reconstruction of TS26 (E17.5) - a collaboration with Glaxo Wellcome Medicines Research Centre, Stevenage.
Dr Jacob Hecksher-Sorensen & Dr James Sharpe, MRC Human Genetics Unit.
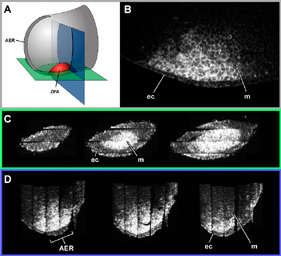
3D Gene Expression - Jacob Hecksher-Sorensen & James Sharpe, 3D Confocal Reconstruction of Gene Expression in the Mouse, Mechanisms of Development (2001).

Dr Chris Armit, Glasgow School of Art.
"Morphogenesis":exploring the 3D animation possibilities using data obtained with the imaging technique Optical Projection Tomography (OPT).
Dr James Sharpe, MRC Human Genetics Unit.

Optical Projection Tomography.