What is EMAP?
The e-Mouse Atlas Project.
EMAP is the combined research projects of Dr Duncan Davidson and Prof Richard Baldock.
Resources provided by EMAP are the EMA Anatomy Atlas of Mouse Development and the EMAGE Gene Expression Database.
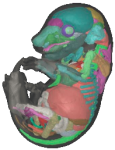
The EMA Anatomy Atlas of Mouse Development uses embryological mouse models to provide a digital atlas of mouse development. It is based on the definitive books of mouse embryonic development by Theiler (1989) and Kaufman (1992) yet extends these studies by creating a series of three dimensional computer models of mouse embryos at successive stages of development with defined anatomical domains linked by a stage-by-stage ontology of anatomical names.
What is EMA?
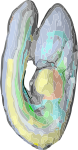
The e-Mouse Atlas (EMA) is a detailed model of the developing mouse. Its purpose is not only to provide information about the shape, gross anatomy and detailed histological structure of the mouse, but also to provide a framework into which information about gene function can be mapped. The spatial, temporal and anatomical integration achieved by mapping diverse data to the organism itself rather than simply to database tables, allows powerful computational analyses. An example of data mapped to the atlas is EMAGE where gene expression patterns can be queried and analysed on a spatial basis.
EMA is an ongoing project. The website presents 3D volumetric models of the embryo linked, in most cases, to comprehensive and detailed images of histological structure. It also contains a detailed anatomy ontology that gives the names and structural relationships between the parts, the 'EMAP anatomy ontology'. The embryo models and anatomy in EMA are organized by standard developmental stage (TS. after Theiler, 1989) and age of the embryo in days post coitum (dpc.).
EMA can be used as an educational tool to teach mammalian developmental biology or as a research tool. The integrated presentation of these resources on this website helps researchers to stage embryos, find anatomical structures, explore detailed histological changes from stage to stage, relate data that is mapped to an EMA embryo model or annotated with terms from the EMAP anatomy ontology to the structure of the embryo and to patterns of gene expression in EMAGE.
What is EMAGE?
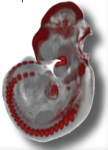
EMAGE is a database of in situ gene expression data in the mouse embryo and an accompanying suite of tools to search and analyse the data.
mRNA in situ hybridisation, protein immunohistochemistry and transgenic reporter data is included. These are sourced from the community and our curators take this data and describe it in a standardised way that allows data query and exchange. The description includes a text-based component but the unique aspect of EMAGE is its spatial annotation focus.
What is the eHistology Kaufman Annotations Resource?
The eHistology Atlas is an online digital resource that allows researchers to interactively explore a collection of cellular-resolution colour histology images detailing mouse development. The haematoxylin and eosin stained sections used in this resource were originally photographed and published in greyscale format in M.H. Kaufman's "The Atlas of Mouse Development" (Kaufman MH (1992) The atlas of mouse development, 1st edn. Academic Press, New York). The original histological sections have been re-digitised at high resolution, in colour and in collaboration with Elsevier, the eHistology Atlas provides the annotations as presented in the original printed version. Only the new section-images are available online, Elsevier plan to use these images to produce an e-book Atlas that will have all of the printed content.
More about the eHistology Histology Atlas.
What is the eLearning Resource?
• E-Learning is a collaboration between Professor José García Monterde of the University of Córdoba and the University of Edinburgh´s eMouseAtlas Project
The aim of this collaboration is:
- to use web technology to deliver Educational Tutorials that enable conceptual understanding of key principles of embryonic development
- to couple the tutorials to High-end 3D Visualisations of mouse developmental anatomy (EMAP) and a database of developmental gene expression (EMAGE)