JAtlasViewer
JAtlasViewer is a program for selecting and viewing
virtual sections from woolz format 3D images. Since any arbitrary section
plane can be chosen, it can be used to explore the internal space of these
3D images. Sample 3D woolz data sets for download:
Data Set Installation: Depending upon which file you down-loaded you can install the sample data-set using:
Microsoft Windows users may prefer to use Winzip. The down-loaded file will unpack to a folder calledcs17 which contains:
Screenshots: 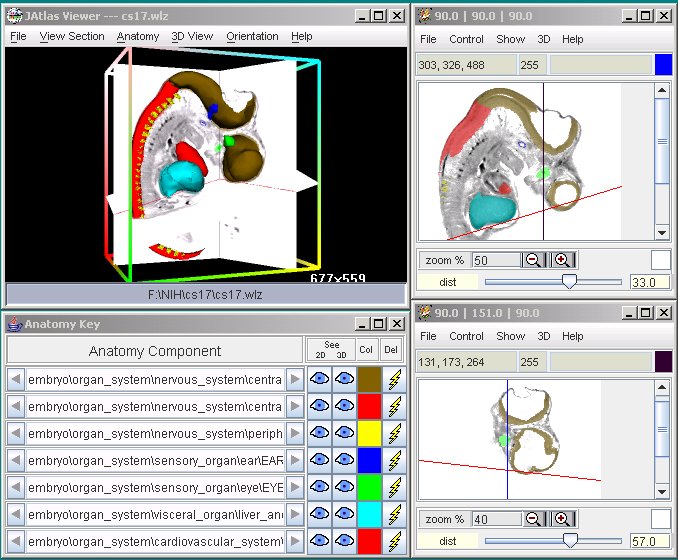
This work has been partially funded
by the NIH HBP grant "Electronic Atlas of the Human Embryonic Brain
Gene Expression" in order to view digital reconstructions of human
and mouse embryos at different stages of development. |