Image Capture
- Summary of configurations
- Configuration (A)
- Configuration (B)
- Configuration (C)
- Configuration (D)
- Configuration (E)
Summary of configurations
A variety of camera and microscope configurations have been used to capture digital images from the prepared microscope slides.
The configurations are summarised in the table.
| Stage | Objective used | Bin | Configuration |
|---|---|---|---|
| ts7 | x10 | A | |
| ts8 | x10 | A | |
| ts9 | x10 | A | |
| ts10 | x10 | A | |
| ts11 | x10 | A | |
| ts12 | x10 | A | |
| ts13 | x10 | A | |
| ts14 | x10 | A | |
| ts15 | x10 | 2 | C |
| ts16 | x10 | 1 | B |
| ts17 | x10 | 2 | B |
| ts18 | x10 | 1 | B |
| ts19 | x10 | 2 | C |
| ts20 | x10 | A | |
| ts23 | x20 | 1 | D |
| ts26 | x2.5 | E |
Configuration (A)
- Microscope : Zeiss Axioplan II
- Lens : 10x neoflour objective
- Camera : Xillix Microimager 1400, 12bit CCD digital
- Software : MAXmgrab software (Unix)
Protocol
Pre-image capture preparation
Sections were serially sectioned as ribbons and laid out on standard 1x3" or 2x3" microscope slides as described in Histology - Epoxy resin.
Digitisation - xmgrab
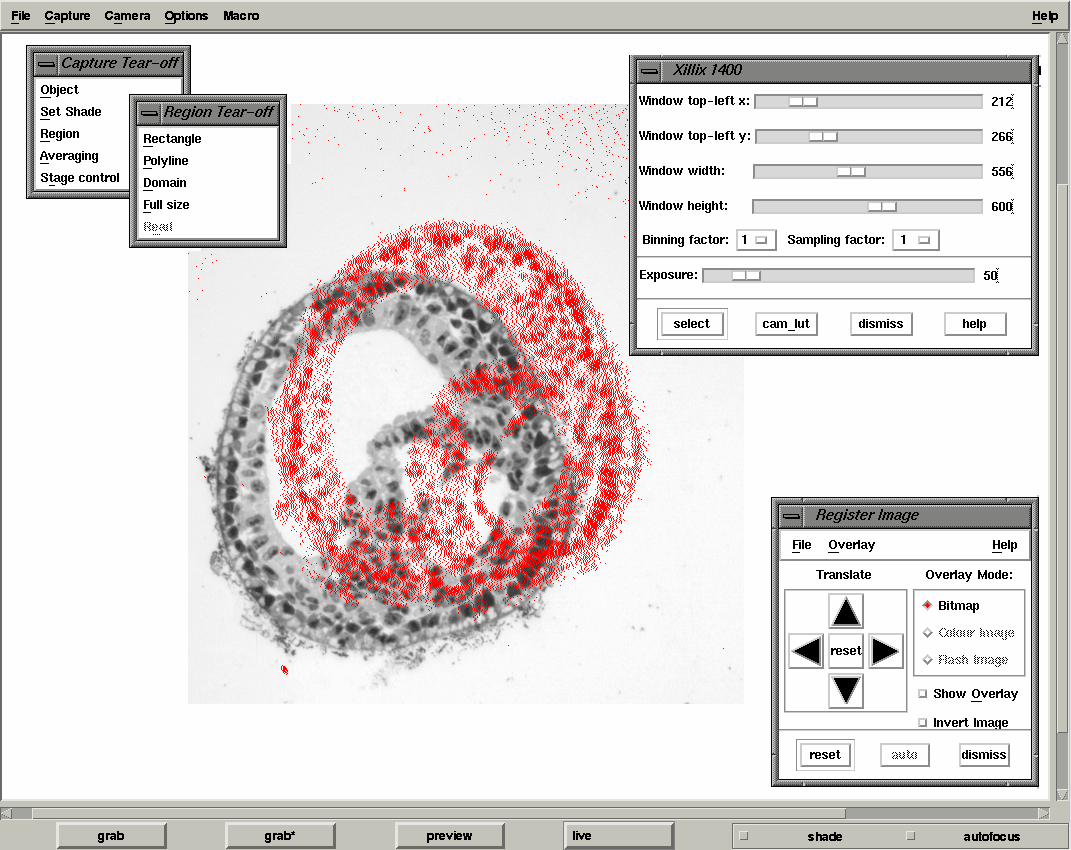
For imaging we use a standard Zeiss Axioplan II microscope with a bright-field condenser specisfically designed for low magnification. For the wax sections we are using a x5 Neoflour objective and for plastic a x10 Neoflour objective. In both cases a green filter was used to enhance contrast. The digitisation is done using a Xillix Microimager 1400 digital CCD camera with the Kodak KAF 1300x1040 chip. This has a square pixel array with a separation between pixels of 6.8 microns. The camera is connected to a Sun Microsystems Sparc-10 workstation via an s-bus interface to capture an image by DMA which can then displayed on the workstation screen. The camera interface, device driver and software libraries for controlling the camera and capturing data have all been developed at the MRC Human Genetics Unit. To provide interactive image capture tuned to the problem of serial sections we have developed the program xmgrab which allows the setting of all camera controls (including the 12-bit to 8-bit LUT), region of interest and provides overlay feedback for rough image registration. In all cases the images are shade-corrected for uneven illumination. xmgrab also allows for the option of averaging a number of frames in order to reduce noise but this has only proved necessary when a standard video camera is used.
The output from xmgrab is a series of 2D image files in woolz format (the MRC HGU image-processing library). The files are now accessed by the program reconstruct which allows review ond correction of the images as well as manual/automatic realignment, grey-level normalisation and rescaling. This is described further in Image Processing - Method 1.
| xmgrab interface showing the alignment overlay |
 |
Configuration (B)
- Microscope : Zeiss Axioplan II
- Lens : 10x neoflour objective
- Camera : Micropublisher 5 Colour
- Software : IPLab (Apple)
Configuration (C)
- Microscope : Zeiss Axioplan II
- Lens : 10x neoflour objective
- Camera : Monochrome QImaging Retiga camera system with 3 colour filters
- Software : IPLab (Apple)
Configuration (D)
- System : Olympus Dotslide
Description
The Olympus DotSlide is an imaging system for 'virtual microscopy', the digital equivalent to conventional light microscopy. Instead of viewing a specimen through the eyepiece of the microscope, a virtual overview image of the entire slide is displayed in perfect quality on the monitor after the system has scanned the specimen at the required resolution. Integrated focus routines make sure the image is always in sharp focus. The single images initially acquired during the scanning process are automatically stitched together to form a large seamless overview image (the 'virtual slide').
| image capture system |
 |
Configuration (E)
- System : Leica Qwin at Glaxo Wellcome Medicines Research Centre, Stevenage.
Protocol
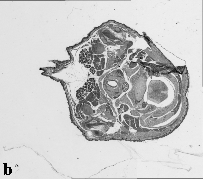
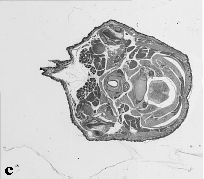
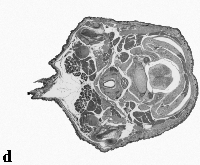
The embryo was captured at a magnification of x2.5 using the Leica Qwin system in Stevenage. Each section is made up of series of up to 12 separate images (Fig. 1a). After going through an automatic patching process the full section image can be seen (Fig. 1b). Due to defects caused by the sectioning process the images require digital correction using graphical software (Fig. 1c). Figure 1d shows a final section. Here the grey level image has been cut out from the background and a random background added (mean = 250, S.D. = 3) using the Mouse Atlas software. This image has been subsampled by 2 to give a final pixel size of 10.4 x 10.4 microns.
| Figure 1: This figure shows the process involved from capturing the digital images (a), to patching them together (b), to digitally repairing them (c) and finally adding a random background to give the final set of sections used in the actual 3D model | 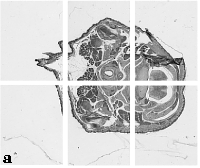 |
 |
 |
 |