Data Sources
The data held in EMAGE comes from three main sources:
|
|
| Literature |
Data from the literature is entered into EMAGE in collaboration with our sister database, the GXD at Mouse Genome Informatics (MGI). GXD curation staff locate mouse gene expression data in the literature and compile a Gene Expression Literature Index which contains information sourced from a very wide range of journals (nearly 600). The Gene Expression Literature Index includes information on the citation, authors, genes/proteins assayed in each paper, whether the samples were whole-mount or sectioned material and the age of the specimens involved.
GXD staff then go on to fully annotate a proportion of this data, by describing in detail the specimen; the detection reagent and methods used and the sites of expression (a text-based description is produced by annotating terms in the EMAP anatomy ontology). This allows query of the GXD database by many aspects.
EMAGE imports some fully annotated data from GXD. EMAGE curators also use the GXD Gene Expression Literature Index to locate data from about 150 journals for full-indexing (including spatial annotation). If a journal does not license its material under a suitable Creative Commons License we have arranged individual legal agreements with the publishers of specific journals (that collectively house over 80% of published in situ gene expression images in the mouse), which allows us to reproduce copyrighted images from these journals on the EMAGE website. Note that if we do not have permission to reproduce the original data image, it is our policy to use a generic image showing the copyright symbol on the EMAGE website that also includes a relevant link to the original data at either PubMed entry or a DOI link direct to the data at the journal website.
Data originating in the literature that is fully annotated in the EMAGE and GXD databases sometimes overlaps and sometimes does not:
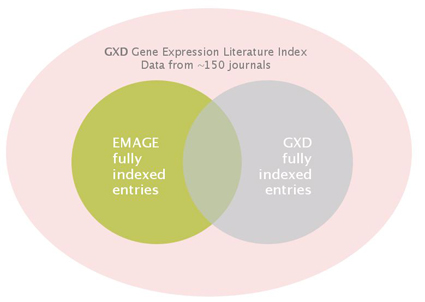
We are committed to better integration of EMAGE and the GXD in the future to produce a resource (MGEIR) that will unify the annotated data.
|
|
|
| Large-scale projects |
EMAGE contains a proportion of data originating from large-scale gene expression screening projects. One current notable example is EURExpress.
The EURExpress consortium generated mRNA in situ hybridisation data for ~20,000 mouse genes on sagittal sections at E14.5 (~24 evenly spaced sections for each gene), and performed a text-based annotation of the sites of expression seen in all 480,000 images. The text annotation was performed manually, with annotation staff visually assessing each image and then using the EMAP anatomy ontology to describe sites of expression.
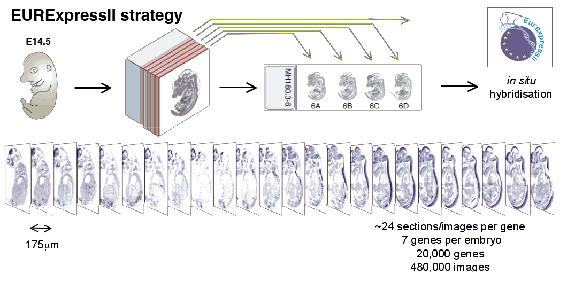
In addition to the information already compilied by the EURExpress consortium, EMAGE developed an automated signal extraction and alignment methods to allow spatial-based annotation and analyses to be applied to this dataset.
Data from other large-scale screens incorporated into EMAGE include:
- Mahoney Transcription Factor data - as published by Gray et al, "Mouse brain organization revealed through direct genome-scale TF expression analysis. " Science. 2004 Dec 24;306(5705):2255-7. This dataset constitutes WM mRNA in situ hybridisation data for ~1350 transcription and other nuclear factors at ~10.5 dpc.
- FaceBase - 3D image data depicting mRNA in situ hybridisation patterns for ~500 genes involved in craniofacial development, at several stages of development. This work was produced in the laboratories of Dr David FitzPatrick (MRC Human Genetics Unit, Edinburgh) and Dr Mike Dixon (School of Dentistry, Manchester University) and was funded as part of the NIDCR P50 DE016215-01 Craniofacial Anomalies Research Center.
- VISTA is a resource of experimentally validated human and mouse non-coding genomic DNA fragments with gene enhancer activity as assessed in transgenic mice. In this project, enhancer candidate sequences were identified by extreme evolutionary sequence conservation or by ChIP-seq. PCR primers were used to amplify conserved regions and ChIP-seq peaks, with the primers chosen extending by several hundred base pairs in both directions to include the flanking sequence required for enhancer activity. The PCR products were then cloned into an Hsp68 coupled LacZ reporter vector and microinjected into fertilized eggs. The embryos were harvested at 11.5 dpc, and stained for LacZ, and the resulting activity patterns annotated.
- EMBRYS is a resource of ~24,500 whole mount gene expression images (~1.5 K genes) from 9.5 dpc, 10.5 dpc and 11.5 dpc mouse embryos. The images generated by the EMBRYS project detail the profiles of transcription factor and transcription factor-related factors in mouse development.
|
|
|
| Individual Labs |
We encourage all mouse embryologists and geneticists to deposit their in situ gene expression data in the EMAGE database
We have received direct data submissions from many labs including those of Brigid Hogan, Janet Rossant, Patrick Tam, Virginia Papaioannou, Carol Wicking, Marianne Bronner, Paula Murphy, Yasuhide Furuta, David FitzPatrick, Mike Dixon and Salvador Martinez. |



 icon to keep this page displayed.)
icon to keep this page displayed.)


