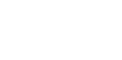Background to the invention
The ability to visualise biological tissues in their correct 3D organisation is of great help to our understanding of biology, and is especially useful in the field of embryo development, in which tissues undergo an intricate array of movements relative to each other. The ability to map the 3D expression patterns of genes and proteins within these complex shapes is also useful, as it gives us insight into their functions, and gives us clues about which genes and proteins interact with each other. With the advent of complete genome sequences, microarray techniques and biological databases, it is likely that the growing need for a comprehensive database of gene expression will become a reality soon. Already a number of laboratories are initiating projects to maps gene expression patterns by the thousand.
The traditional method for creating 3D reconstructions of embryonic tissue is a laborious, time-consuming job, as it involves embedding the tissue, cutting it into hundreds of serial sections, mounting the sections, staining them, digitising them under a microscope, feeding them into an appropriate computer program and finally manually compensating for the distortions which have inevitably been introduced during the whole process.
Much effort has recently been put into the development of techniques which can image the structure of biological tissue in 3D, and in particular the mapping of gene expression patterns. By far the most commonly used is confocal microscopy. Typically, this is used to scan up to a depth of a few hundred microns, although the advent of dual-photon and multi-photon techniques means that good resolution images can be captured at greater depth. For some specimen which are larger than this depth, gene expression patterns can be built-up by scanning serial thick sections.
Although mMRI holds much promise for imaging experiments in the future, especially with its ability to image living specimen, it also displays some limitations. We have developed a new imaging technique which has the following advantages over mMRI:
- It can image coloured stains. This allows us to image data from:
- Normal in-situ hybridisation experiments (using the BCIP and NBT substrates)
- Reporter gene experiments (such as LacZ and alkaline phosphatase)
- It can image fluorescent dyes. This allows us to image:
- Multiple signals from the same tissue
- Standard fluorescent antibody reagents
- Only takes about 15 minutes to image each channel. This speed allows us to image many specimen rapidly, and therefore to perform studies of natural variation in shape, gene expression, mutant phenotypes etc.
- It is much cheaper than mMRI, and works in conjunction with a normal "dissecting" microscope, making it accessible to many more biologists.